What if researchers could go back in time 105 million years and accurately sequence the chromosomes of the first placental mammal? What would it reveal about evolution and modern mammals, including humans?
In a study published this week in Proceedings of the National Academy of Sciences, researchers have gone back in time, at least virtually, computationally recreating the chromosomes of the first eutherian mammal, the long-extinct, shrewlike ancestor of all placental mammals.

“The revolution in DNA sequencing has provided us with enough chromosome-scale genome assemblies to permit the computational reconstruction of the eutherian ancestor, as well as other key ancestors along the lineage leading to modern humans,” said Harris Lewin, a lead author of the study and a professor of evolution and ecology and Robert and Rosabel Osborne Endowed Chair at the University of California, Davis.
“We now understand the major steps of chromosomal evolution that led to the genome organization of more than half the existing orders of mammals. These studies will allow us to determine the role of chromosome rearrangements in the formation of new mammal species and how such rearrangements result in adaptive changes that are specific to the different mammalian lineages,” said Lewin.
The findings also have broad implications for understanding how chromosomal rearrangements over millions of years may contribute to human diseases, such as cancer.
“By gaining a better understanding of the relationship between evolutionary breakpoints and cancer breakpoints, the essential molecular features of chromosomes that lead to their instability can be revealed,” said Lewin. “Our studies can be extended to the early detection of cancer by identifying diagnostic chromosome rearrangements in humans and other animals, and possibly novel targets for personalized therapy.”
Descrambling chromosomes
To recreate the chromosomes of these ancient relatives, the team began with the sequenced genomes of 19 existing placental mammals — all eutherian descendants — including human, goat, dog, orangutan, cattle, mouse and chimpanzee, among others.
The researchers then utilized a new algorithm they developed called DESCHRAMBLER. The algorithm computed (“descrambled”) the most likely order and orientation of 2,404 chromosome fragments that were common among the 19 placental mammals’ genomes.
“It is the largest and most comprehensive such analysis performed to date, and DESCHRAMBLER was shown to produce highly accurate reconstructions using data simulation and by benchmarking it against other reconstruction tools,” said Jian Ma, the study’s co-senior author and an associate professor of computational biology at Carnegie Mellon University in Pittsburgh.
In addition to the eutherian ancestor, reconstructions were made for the six other ancestral genomes on the human evolutionary tree: boreoeutherian, euarchontoglires, simian (primates), catarrhini (Old World monkeys), great apes and human-chimpanzee. The reconstructions give a detailed picture of the various chromosomal changes — translocations, inversions, fissions and other complex rearrangements — that have occurred over the 105 million years between the first mammal and Homo sapiens.
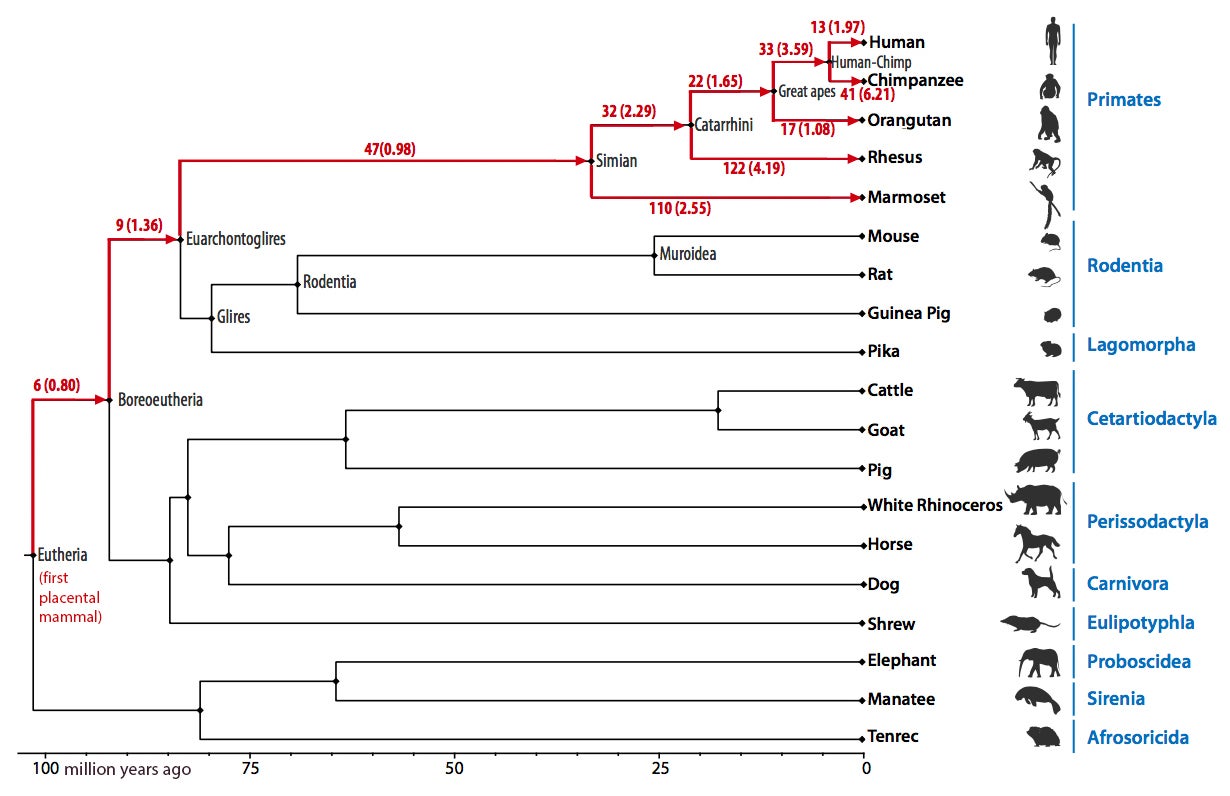
Rates of evolution vary
One discovery is that the first eutherian ancestor likely had 42 chromosomes, four less than humans. Researchers identified 162 chromosomal breakpoints — locations where a chromosome broke open, allowing for rearrangements — between the eutherian ancestor and the formation of humans as a species.
The rates of evolution of ancestral chromosomes differed greatly among the different mammal lineages. But some chromosomes remained extremely stable over time. For example, six of the reconstructed eutherian ancestral chromosomes showed no rearrangements for almost 100 million years until the appearance of the common ancestor of human and chimpanzee.
Orangutan chromosomes were found to be the slowest evolving of all primates and still retain eight chromosomes that have not changed much with respect to gene order orientation as compared with the eutherian ancestor. In contrast, the lineage leading to chimpanzees had the highest rate of chromosome rearrangements among primates.
“When chromosomes rearrange, new genes and regulatory elements may form that alter the regulation of expression of hundreds of genes, or more. At least some of these events may be responsible for the major phenotypic differences we observe between the mammal orders,” said Denis Larkin, co-senior author of the study and a reader in comparative genomics at the Royal Veterinary College at the University of London.
The chromosomes of the oldest three ancestors (eutherian, boreoeutherian, and euarchontoglires) were each found to include more than 80 percent of the entire length of the human genome, the most detailed reconstructions reported to date. The reconstructed chromosomes of the most recent common ancestor of simians, catarrhini, great apes, and humans and chimpanzees included more than 90 percent of human genome sequence, providing a structural framework for understanding primate evolution.
Collaborators and funding
Collaborating with Lewin, Larkin and Ma were Marta Farré of the Royal Veterinary College at the University of London in London, England; Jaebum Kim from the Department of Biomedical Science and Engineering at Konkuk University in Seoul, South Korea; and Loretta Auvil and Boris Capitanu of the Illinois Informatics Institute at the University of Illinois at Urbana–Champaign in Urbana, Illinois.
The research was supported by the Biotechnology and Biological Sciences Research Council; the Robert and Rosabel Osborne Endowment (UC Davis); the National Institutes of Health; the National Science Foundation; the Ministry of Science, ICT & Future Planning of Korea; the Ministry of Education of Korea; and the Rural Development Administration of Korea. This work was conducted as a contribution to the Genome 10K Project.
Media Resources
Harris Lewin, UC Davis Evolution and Ecology, lewin@ucdavis.edu
Andy Fell, UC Davis News and Media Relations, 530-752-4533, ahfell@ucdavis.edu
